
Adjusting the pH below or above the protein pI induces a positive charge when pH pI (Fig.1).įig.1: Protein charge is function of Buffer pH Consequently, the protein is not able to bind to an ion exchange column (anion and cation). Indeed, when the pH buffer is equal to the pI (isoelectric point), the protein has no charge. For example, if you’re planning an ion exchange purification you need to choose the right pH to have your protein charged as you want. The first thing to do is to determine at which pH you need to work.
Compatibility with subsequent experiments. Compatibility with other components of the buffer. Obviously, it is not so simple and it will require some optimization, but I think that a “what-to-do-first-guide” is always welcome! 1#Definitionįirst of all, a buffer solution is one which resists changes in pH when small quantities of an acid or an alkali are added to it.Ī good buffer needs to have some specific characteristics like: I’d like to offer you this simple guide, which will allow you to easily select the right basic buffer for the right protein purification strategy in order to facilitate your decision making. You will be faced with a bewildering array of choices leading you to ask yourself “where do I start…?”. Your ultimate aim is to keep your protein “happy” by choosing the perfect buffer. Indeed, the quest does not stop there! The next step is purification, during which you will try to isolate your protein of interest from the surrounding contaminants while keeping it soluble and active. Once you have successfully achieved the production of your protein in a selected system, you need to think about the following steps. In general, the pI values for native proteins are affected by the three-dimensional structure of the proteins, which causes greater differences between calculated and experimental pI values than in the case of polypeptides for which pI values are determined in the presence of urea.Protein biochemistry brings together a vast and varied world of methods of protein production, purification and characterization. The pI of the native Glut1 was lower, 8.0 +/- 0.1, at 22 degrees C. The calculated pI for the human red cell glucose transporter (Glut1) with one sialic acid residue was decreased from 8.8 to 8.5 by introducing pKa value spreading and became consistent with the experimental pI value of 8.4 +/- 0.05 at 15 degrees C determined in the presence of 6 M urea. The calculated pI values showed reasonably good agreement with experimental ones for most of 16 native proteins over a wide pH range (3.4-11) when charge contributions of heme groups, sialic acid residues, etc., were taken into account. Each particular type of ionizable group was assumed to have pKa values distributed around the chosen value, thereby simulating the situation in proteins and polypeptides. 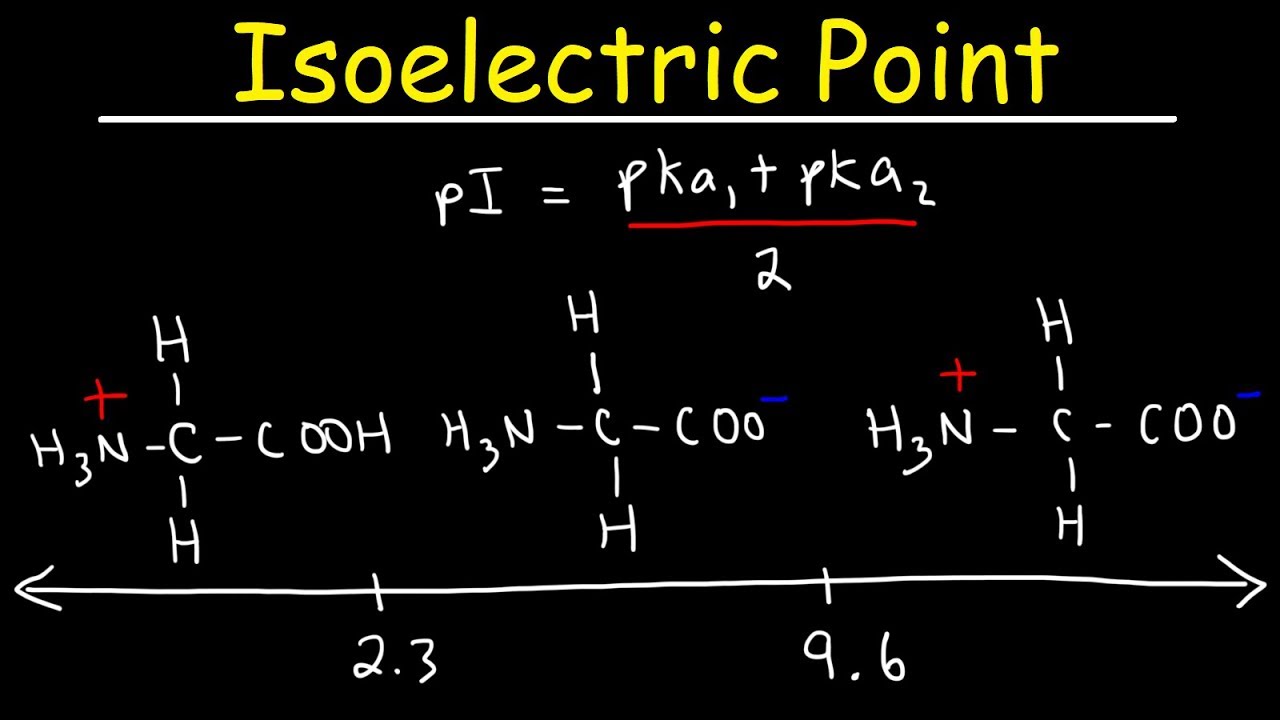
A set of pKa values was chosen for amino acid residues with ionizable side chains. Amino acid composition, pKa values for amino acid side chains and for the N- and C-terminal groups, and the presence of other charged groups were taken into account.
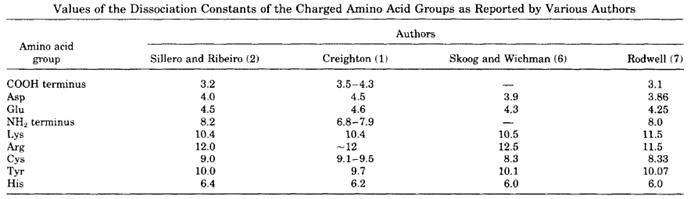
For estimating pI values the net charge of several proteins was calculated versus pH by use of the Henderson-Hasselbalch equation.
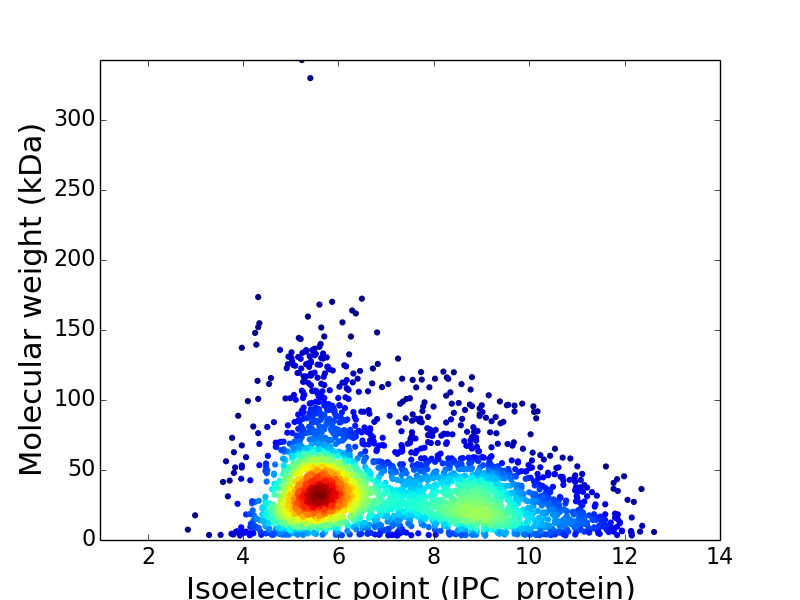
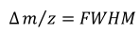
The isoelectric points (pI) of native proteins are important in several separation techniques.







 0 kommentar(er)
0 kommentar(er)
